
Develop Tool to Improve CRISPR Off-Target Predictions Using Genetic Variants
Researchers develop a web-based tool that improves CRISPR accuracy by identifying off-target effects across genetic variations
CRISPR-based gene editing holds great promise, but off-target effects remain a major concern, especially across diverse genetic backgrounds. A new study presents a web-based tool that enhances off-target site prediction by incorporating individual genetic variants. Developed using the human genome and pepper plant cultivars, the tool improves accuracy at the haplotype level. This user-friendly, login-free platform offers researchers a powerful way to personalize and safeguard genome editing applications across fields.
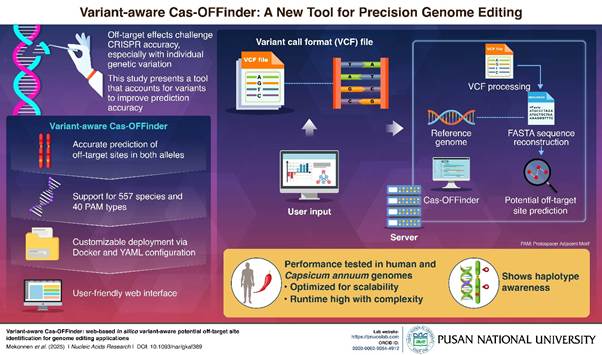
Image title: New Tool Enhances CRISPR Accuracy by Accounting for Genetic Variants
Image caption: Researchers introduce Variant-aware Cas-OFFinder, a web-based tool that improves CRISPR off-target predictions by incorporating individual genetic differences. By reconstructing allele-specific sequences, the tool offers haplotype-level insights, enabling safer and more precise genome editing in medicine, agriculture, and research.
Image credit: Professor Jeongbin Park from Pusan National University, Korea
License type: Original Content
Usage restrictions: Cannot be reused without permission
Genome editing technologies like CRISPR-Cas9 have transformed biology, medicine, and agriculture, but concerns remain about unintended edits at off-target DNA sites. These off-target effects can cause harmful mutations and are difficult to predict, as they depend not only on the guide RNA but also on an organism’s unique genetic makeup. Most existing prediction tools rely on a standard reference genome, ignoring key variations like SNPs, insertions, and deletions that differ across individuals and alleles.
To address this issue, researchers led by Professor Jeongbin Park from Pusan National University developed Variant-aware Cas-OFFinder, a web-based tool that improves off-target site prediction by accounting for individual genetic variation. Co-first authors Abyot Melkamu Mekonnen (PhD candidate) and Kang Seong (undergraduate at the time, now a master’s student at KAIST), who conducted the study while at Pusan National University, said, “It is very rewarding to know that the technology we developed can be of practical help in correcting individual genes.” The tool was tested using the human genome and pepper cultivars. Their findings were published online on 8 May, 2025 in the journal Nucleic Acids Research.
The Variant-aware Cas-OFFinder tool offers a significant step forward in personalized genome editing by incorporating genetic diversity directly into off-target predictions. It accepts phased single-sample VCF files and supports GPU acceleration, allowing application beyond standard model organisms. By reconstructing allele-specific genome sequences, the tool performs haplotype-level analysis, offering a detailed and accurate picture of potential off-target effects.
“In my view, genome editing tools should be as individualized as the genomes they target,” says Prof. Park. “Our tool lays the groundwork for more accurate and personalized editing strategies.”
The team tested the tool on human and sweet pepper (Capsicum annuum) genomes using public and cultivar-specific sequencing data. In humans, it revealed off-target sites on chromosome 10 absent from the standard reference genome. In sweet pepper, it identified distinct allele-specific off-targets, useful for plant breeding.
Compared with existing tools like Cas-OFFinder and CRISPRitz, Variant-aware Cas-OFFinder consistently identified unique off-targets caused by small variants such as insertions and deletions. Although it does not yet detect large structural variants, it supports 557 species and 40 PAM types, and is fully customizable through YAML files.
“Precision genome editing requires precision tools,” remarks Prof. Park. “We believe Variant-aware Cas-OFFinder will play a central role in developing safer, more effective CRISPR therapies and agricultural applications.”
The tool is available both as a user-friendly web interface and a command-line version. Its source code, benchmark tools, and example datasets are freely available on GitHub and Zenodo. While its haplotype-level analysis can slow performance slightly compared to earlier tools, this trade-off enables greater accuracy and reliability in genome editing research.
Reference
Title of original paper: Variant-aware Cas-OFFinder: web-based in silico variant-aware potential off-target site identification for genome editing applications
Journal: Nucleic Acids Research
DOI: 10.1093/nar/gkaf389
About Pusan National University
Pusan National University, located in Busan, South Korea, was founded in 1946 and is now the No. 1 national university of South Korea in research and educational competency. The multi-campus university also has other smaller campuses in Yangsan, Miryang, and Ami. The university prides itself on the principles of truth, freedom, and service and has approximately 30,000 students, 1,200 professors, and 750 faculty members. The university comprises 14 colleges (schools) and one independent division, with 103 departments in all.
Website: https://www.pusan.ac.kr/eng/Main.do
About Professor Jeongbin Park
Prof. Jeongbin Park is an Assistant Professor leading the Computational Omics Laboratory at Pusan National University since 2022. He holds degrees in Physics (BSc, Pusan National University, 2012; MSc, Seoul National University, 2014) and Biology (PhD, Heidelberg University, 2020). His research focuses on computational oncology and spatial omics data analysis. He has held positions at the German Cancer Research Center, Berlin Institute of Health, and Institute for Basic Science. Prof. Park is known as the lead developer of CRISPR RGEN Tools, a widely used genome editing platform.
Lab:
ORCID ID:0000-0002-9064-4912

 PURCS_154_Infographic_final.jpg
(1MB)
PURCS_154_Infographic_final.jpg
(1MB)